Oligo Database Functionality 1
|
![]()
Oligo Database Functionality 1
|
![]()
The Database window allows you to create and save new databases for storing oligonucleotide and oligonucleotide sets data. It is also a powerful analysis and multiplex search tool. After any Oligo search you may import all or some found oligos to the database.
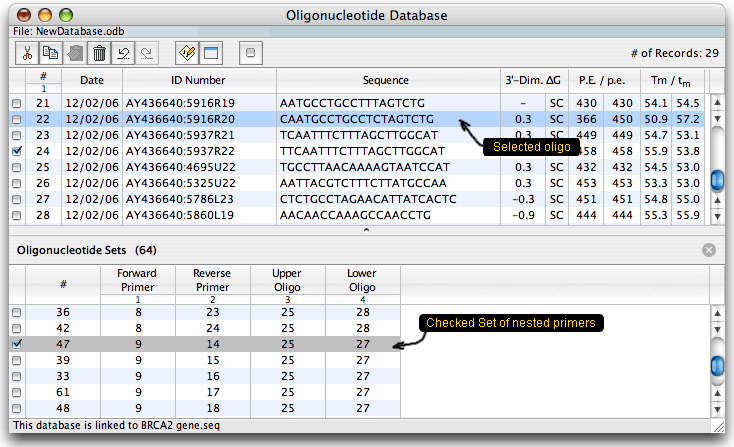
The database is displayed as a table. Columns on the right show certain analysis data for each oligo: the strongest dimer free energy, compatibility (SC means self-compatible), priming efficiency (P.E.) and Tm. When the database is linked to a sequence, it shows absolute P.E. and Tm values and relative to the linked sequence values that consider mismatches (p.e. and tm). You may check or select oligos or oligo sets and then analyze them for dimers & hairpins or export to another sequence. You may also run a search to find all cross-compatible primer sets (multiplex database). After the search, when you choose a group of cross-compatible primers and click on the Select button you'd get this:
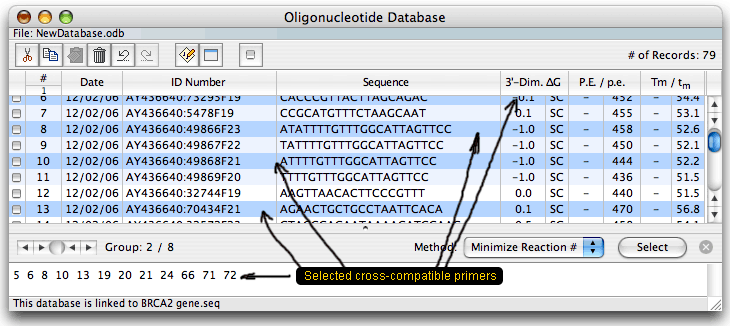
The primers of one group should not interfere with each other in one reaction mixture. The database has its own parameters, so unlike the previous Oligo version, you may open it and analyze the oligos without opening the parent sequence file. When this is done, you won't be able to calculate the real P.E. and Tm - you'd see just the dashes as shown above.
The most versatile and time saving option is multiplexing entire sets. This is explained on the next page.